ACM KDD-Cup
2004: Tied for 1st Place Overall
INTRODUCTOIN
KDD Cup is the worldwide Data Mining and Knowledge Discovery competition organized by ACM SIGKDD (Special Interest Group on Knowledge Discovery and Data Mining). KDD Cup is held annually in conjunction with the ACM SIGKDD conferences. It aims at showing the best methods for discovering higher-level knowledge from data, helping to close the gap between research and industry and stimulating further KDD research and development.
KDD Cup 2004 includes two tasks, particle physics task and protein homology prediction task. The goal is to optimize learning for different performance metrics. This is the first time we have participated the KDD Cup competition and we won the tied for the 1st place overall and honorable mentions for Squared Error and Average Precision metrics in the protein homology task.
TEAM MEMBERS
Yan Fu, Ruixiang Sun, Qiang Yang, Simin He, Chunli Wang, Haipeng Wang, Shiguang Shan, Junfa Liu, Wen Gao
TASK DESCRIPTION
The goal of protein homology prediction task in KDD Cup 2004 is to predict which proteins in database are homologous to a native (query) sequence. Each database protein sequence is described by 74 features that measure the similarity between the database protein sequence and the query sequence.
Biological background: Protein structure play an important role in biological functions. Experimental approach to protein structure determination is both slow and time-consuming. Protein homology prediction is a key step of protein structure prediction. Since homologous proteins (evolved from the same ancestor) usually share similar structures, predicting protein structures based protein homology has been one of the most important problems in bioinformatics. The objective of the problem is to find those proteins in the database that are homologous to the query protein so that the homologous proteins can be used as structural templates.
WWW search analogy: “If you are not familiar with protein matching and structure prediction, it might be helpful to think of this as a WWW search engine problem. There are 153 queries in the train set, and 150 queries in the test set. For each query there are about 1000 returned documents, only a few of the documents are correct matches for each query. The goal is to predict which of the 1000 documents best match the query based on 74 attributes that measure match. A good set of match predictions will rank the "homologous" documents near the top.” (from KDD Cup 2004 website)
APPROACH
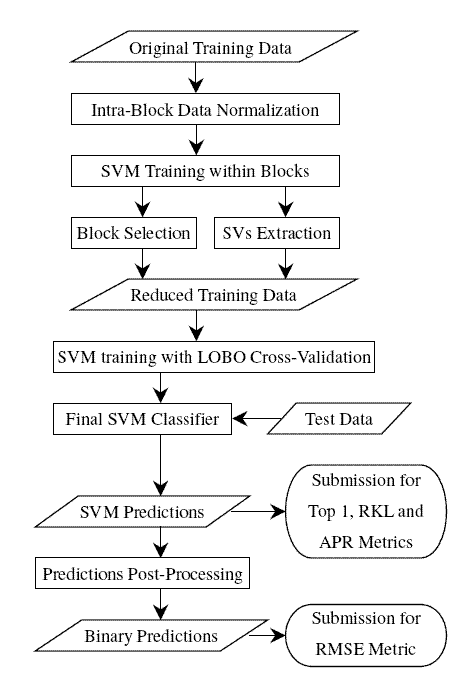
The Protein homology prediction task in KDD Cup 2004 is a machine learning, or specifically, learning to rank problem. A major characteristic of the learning to rank problem is that each feature vector is computed based on a query. Therefore, all feature vectors are partitioned into groups by queries. Each group of data associated with a query is called a block. We developed several block-based or query-dependent approaches to more accurate learning to rank and applies these approaches to the protein homology prediction problem. Below is a flowchart of our winning approaches (see our report on SIGKDD Explorations for details). Currently, we are still working on this problem (a recent technical report).
CERTIFICATE

Previous
KDD Cup
n
KDD Cup 2004, features tasks in
particle physics and bioinformatics evaluated on a variety of different
measures
n
KDD Cup 2003, focuses
on problems motivated by network mining and the analysis of usage logs
n
KDD Cup 2002, focus:
bioinformatics and text mining
n
KDD Cup 2001,
focus: bioinformatics and drug discovery
n
KDD Cup 2000,
focus: web mining tasks
n
KDD Cup 1999,
focus: intrusion detection and report
n
KDD Cup 1998,
focus: direct marketing, list with best donation value, best report
n
KDD Cup 1997,
focus: predicting most likely donors for a charity
You can also visit the following websites to know more about the competition in each year:
http://www.sigkdd.org/kddcup/index.php
http://www.kdnuggets.com/datasets/kddcup.html
Related
News on Our Results
Graduate School of Chinese Academy of Sciences