iPRG
Introduction to the ABRF iPRG2008
Study
A significant challenge in proteome
informatics is accurate and concise reporting of protein identification data
that result from mass spectrometry-based proteomic workflows. The Paris Guidelines
represent the proteomics communityกฏs
efforts to devise a standard methodology for reporting protein identification
data.
Given these guidelines, the Proteome
Informatics Research Group (iPRG) of the Association of Biomolecular Resource
Facilities (ABRF) proposes to assess the consistency of protein identification
analysis. The iPRG2008 study focuses on evaluating the ability of proteomics
laboratories to determine the identities of a complex mixture of proteins present
in a single mass spectral dataset.
The primary goals of this study are
to provide each participating laboratory an opportunity to evaluate its capabilities
and approaches with regard to:
· Bioinformatics tools used to make and consolidate
protein identifications derived from a common data set and a common reference
database.
· Report those identifications using common reporting
criteria.
The data set for this study consists
of mouse samples that were digested with trypsin, resolved into 13 fractions
by strong cation exchange chromatography, and then analyzed by LC-MS/MS (3200
QTRAP). Most of the fractions were analyzed with successive rounds of exclusion
in order to identify more peptides in each fraction. This resulted in the total
of 29 acquisition sets and 41,977 spectra.
The determination of the "correct"
proteins is performed by voting strategy. Each iPRG expert analyzes the data
independently with the tools of their choosing like any study respondent. The
resulted protein identifications are evaluated and graded into several categories
with different confidence. This pool of protein identifications serves as a
benchmark to evaluate the performance of the identification results each participant
submitted.
In January 2008, we participated
this study with pFind (a database-searching software for protein identification
via tandem mass spectrometry) and pCompare (an in-house software tool for peptide
identification result parsing and protein inferring) developed by ourselves.
The performance of our results is shown in the following figures extracted from
an ABRF poster. Now we are taking our efforts to make pFind more reliable and
applicable.
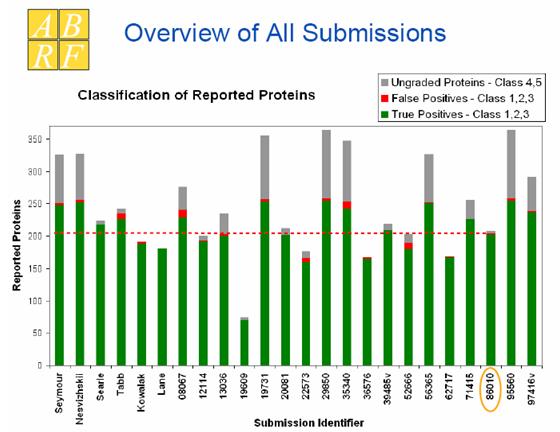
Figure 1. Overview of all
submissions. Each histogram bin represents an iPRG expert (6 bins on the left)
or a participant. The area of histogram bins in green, red, and gray represent
the number of correct, false, and uncertain identifications, respectively. This
analysis shows that the performance of our software (labeled with 86010) is
comparative to that of the experts on an average.
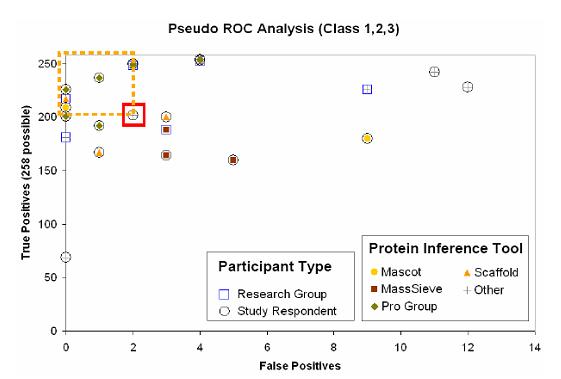
Figure 2. ROC analysis of
all submissions. The most ideal performance should be at the utmostly top-left
corner of the coordinate chart. The dot within the red square represents our
submitted results.
Details about the ABRF iPRG2008
Study, can be found here.

