pFind Studio: a computational solution for mass spectrometry-based proteomics
Introduction
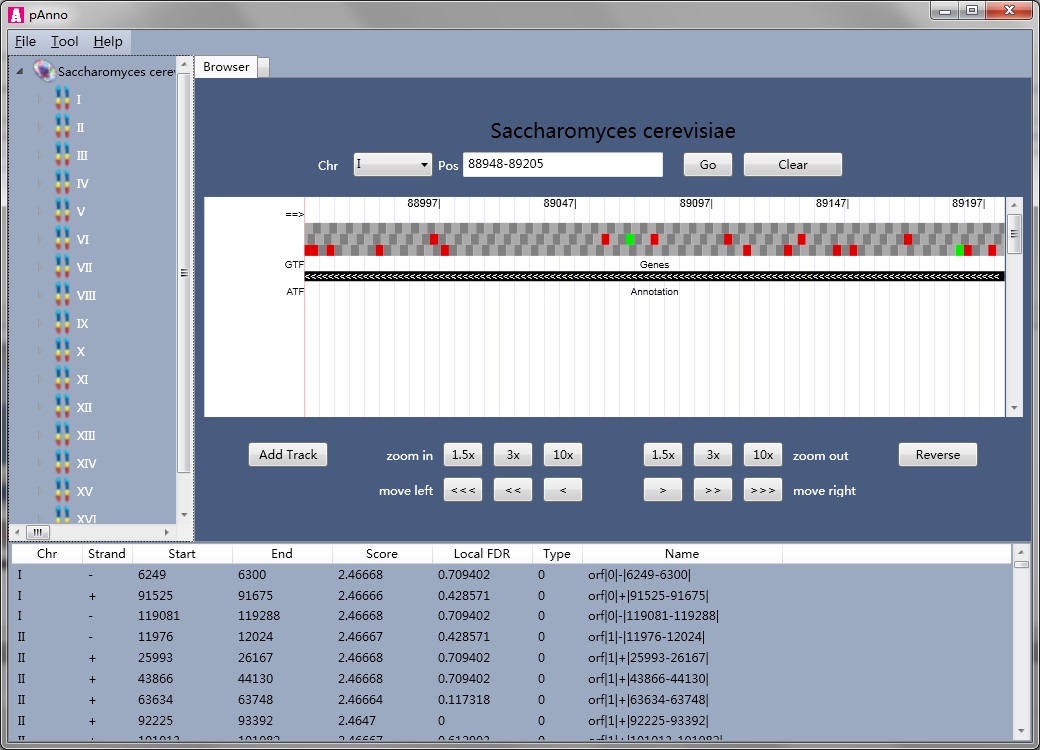
pAnno® is a proteogenomic tool that combines the following functions, (i) browsing genome, (ii) constructing a 6 frame translation protein database for MS/MS identification, (iii) using MS/MS searching result to re-annotate genome.
Database searching-based computational proteomics rely heavily on the completeness and accuracy of the protein sequences, but the protein sequences are not perfect (protein sequences are missing, translation initiation sites are mis-annotated), under which situation we need protegenomic searching. The imperfectness of the protein sequence could be refined by the original genome. Mapping MS/MS spectra to a protein database derived from 6 frame translation to some extent, could be a good solution. However, too much attention should be paid when using this technique, that is, novel PSMs may be false positives. To overcome this problem, we integrate a ORF (generated by 6 frame translation) level scoring strategy into pAnno, and give each ORF a local FDR value. This value could guide users to do further validations.
Cite us
1. A Note on the False Discovery Rate of Novel Peptides in Proteogenomics.
Kun Zhang, Yan Fu, Wen-Feng Zeng, Kun He, Hao Chi, Chao Liu, Yan-Chang Li, Yuan Gao, Ping Xu, Si-Min He.Bioinformatics, 31(20), 3249-3253, 2015. [abstract]
Notice: please quote these above publications when you use pAnno software in your papers.Downloads
pAnno v1.0 is currently free to use. Please read carefully the pAnno Software License Agreement before downloading and using the software. Please fill in the registered table and email it to panno@ict.ac.cn to get the download link.
If you have any questions about it, please contact panno@ict.ac.cn.
Version history
| Release version | Release date | Click for more | |
|---|---|---|---|
| pAnno v1.0 | 2017-12-29 | Download | Release Notes |
 ®Copyright © pFind Team · Institute of Computing Technology, Chinese Academy of Sciences, Beijing, China
®Copyright © pFind Team · Institute of Computing Technology, Chinese Academy of Sciences, Beijing, China