pFind Studio: a computational solution for mass spectrometry-based proteomics
Introduction
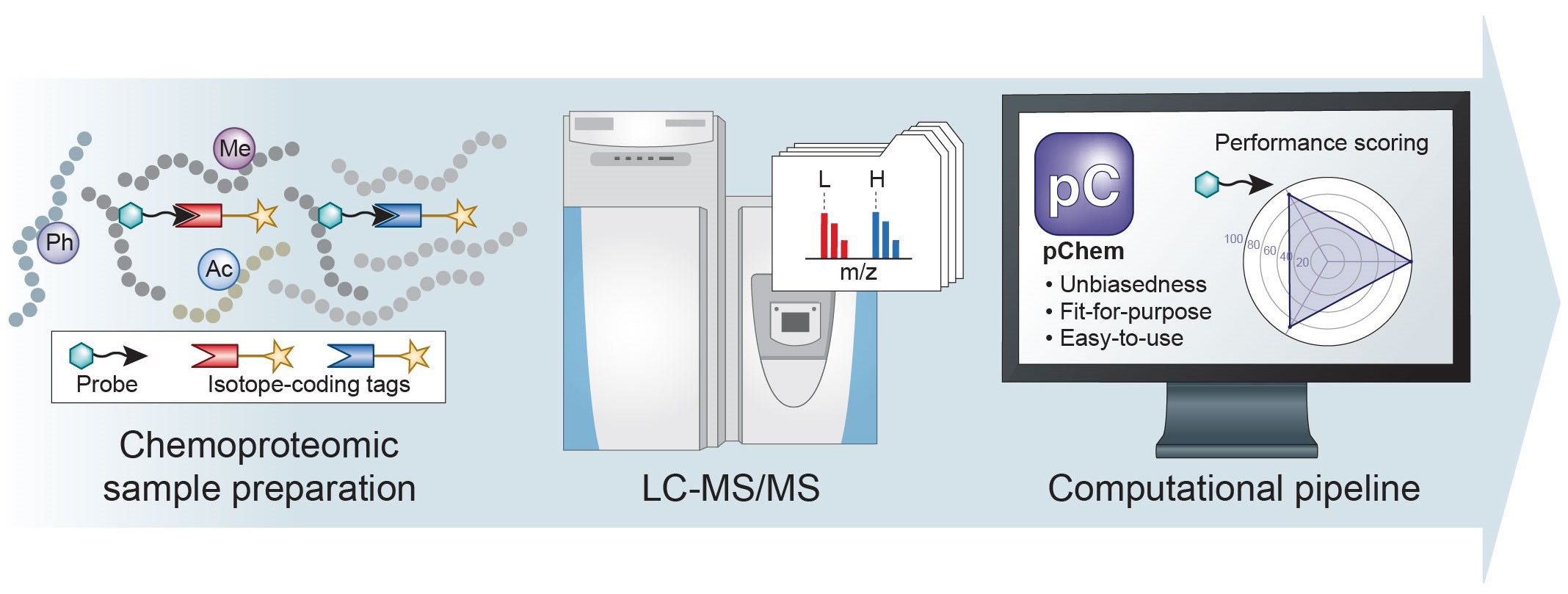
Chemical probe coupled with mass spectrometry (MS)-based proteomics, herein termed chemoproteomics, offers versatile tools to globally profile protein features and to systematically interrogate the mode of action of small molecules in a native biological system. Nonetheless, development of an efficient and selective probe for chemoproteomics can still be challenging. Besides, it is also difficult to unbiasedly assess its chemoselectivity at a proteome-wide scale. Here we present pChem, a modification-centric blind search and summarization tool to provide a pipeline for rapid and unbiased assessing of the performance of ABPP and metabolic labeling probes. This pipeline starts experimentally by isotopic coding of PDMs, which can be automatically recognized, paired, and accurately reported by pChem, further allowing users to score the profiling efficiency, modification-homogeneity and proteome-wide residue selectivity of a chemoproteomic probe.
Cite us
pChem: a modification-centric assessment tool for performance of chemoproteomic probes.
Ji-Xiang He*, Zheng-Cong Fei*, Ling Fu, Cai-Ping Tian, Fu-Chu He, Hao Chi, Jing Yang.Nat Chem Biol (2022).
https://doi.org/10.1038/s41589-022-01074-8
Downloads
pIon is currently available for free use.
Notice: Sep. 29, 2024 - The functionality of pChem 1.1 is also fully supported in this version. Click to download.
For source code, please refer to github.
For detailed usage, please refer to user guide.
pChem version 1.1 is currently free to use.
Notice: Jan. 10, 2023 - If you are using a version prior to this date, please re-download the pChem software in time. The expiration date is set on Jan. 10, 2026.
Please visit i.pfind.org to register and download pChem.
For source code, please refer to github.
For detailed usage, please refer to user guide.
For other issues, please contact pengyaping21s@ict.ac.cn.