pFind Studio: a computational solution for mass spectrometry-based proteomics
Introduction
pFind® 3 is developed as an upgrade of pFind 2.8 (visit the new version).
pFind® is a search engine system for automated peptide and protein identification from tandem mass spectra.
Although many available database searching tools have been developed, there are still a lot of challenges in identification reliability, sensitivity and usability. For example, the target-decoy database strategy has been widely used for the estimation of false discovery rate (FDR) of peptide identification. However, this is usually done manually by users and all of existing tools lack an automated module to estimate the FDR. Another problem is the speed of searching high-throughput spectra against huge peptide and protein databases. The improvements in the sensitivity of mass spectrometers and the rapid expansion of databases have increased the scope and complexity of searching. Traditional software architecture, i.e., running all tasks in a stand-alone process and having not any data index, is more and more inadequate.
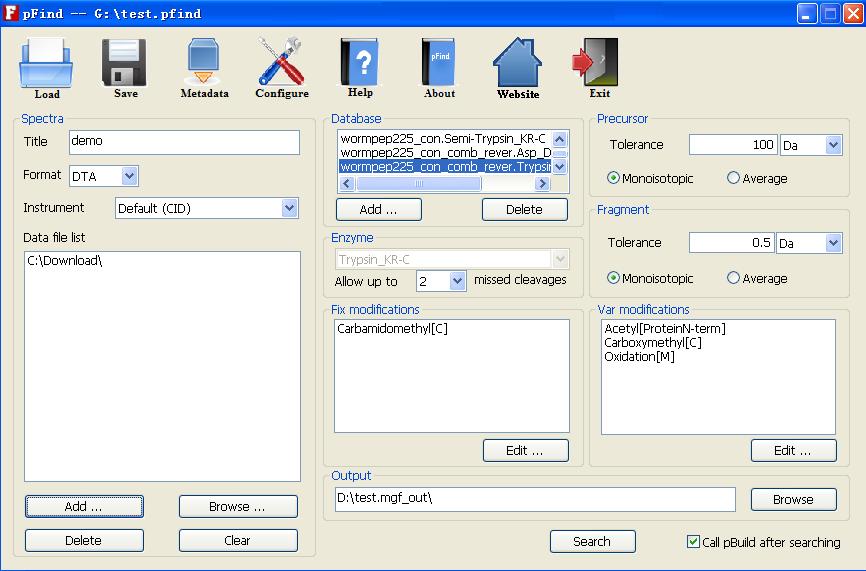
The newest version of pFind incorporate several newly developed or improved algorithms, modules and workflows: The system incorporates the target-decoy database search strategy for automated FPR estimation. Users only need to specify a required FDR before searching. Then the system will calculate a threshold that achieves the FDR and filter search results automatically. We developed a toolbox to index protein databases for high-throughput application and designed all modules under a parallel-processing-oriented architecture for distributing the computational load efficiently among a lot of computers. These developments greatly improve the overall searching speed.
References
Rapid Communications in Mass Spectrometry, 21,2985-2991,2007. [abstract]
pFind 2.0: a software package for peptide and protein identification via tandem mass spectrometry.
Le-Heng Wang, De-Quan Li, Yan Fu, Hai-Peng Wang, Jing-Fen Zhang, Zuo-Fei Yuan,Rui-Xiang Sun, Rong Zeng, Si-Min He, Wen Gao.
Bioinformatics 20, 1948-1954, 2004. [abstract]
Exploiting the kernel trick to correlate fragment ions for peptide identification via tandem mass spectrometry.
Yan Fu, Qiang Yang, Rui-Xiang Sun, De-Quan Li, Rong Zeng, Charles X. Ling, Wen Gao.
Notice: please quote these above publications when you use pFind software in your papers.Downloads
Please read carefully the pFind Software License Agreement before downloading and using the software. Please fill in the registered table and email it to pfind@ict.ac.cn to get the registration key.
pFind v2.8 is currently free to use. Download pFind 2.8.8 (Released on Jan.5, 2018).
If you have any questions about it, please contact pfind@ict.ac.cn.
User guide: How to use pFind
Version history
| Release version | Release date |
|---|---|
| pFind v2.8.8 | 2018-01-05 |
| pFind v2.8 beta1 | 2012-09-14 |
| pFind v2.6 final release | 2011-09-01 |
| pFind 2.3 final release (pFind 2.4) | 2010-04-15 |
| pFind 2.2 final release | 2009-08-05 |
| pFind 2.1 final release | 2008-08-08 |
| pFind 2.0 final release | 2008-02-29 |
| pFind 1.6 final release | 2005-06-01 |
| pFind 1.5 final release (the first indexing version) | 2004-10-01 |
| pFind 1.0 final release | 2003-10-01 |
 ®Copyright © pFind Team · Institute of Computing Technology, Chinese Academy of Sciences, Beijing, China
®Copyright © pFind Team · Institute of Computing Technology, Chinese Academy of Sciences, Beijing, China