pFind Studio: a computational solution for mass spectrometry-based proteomics
Introduction
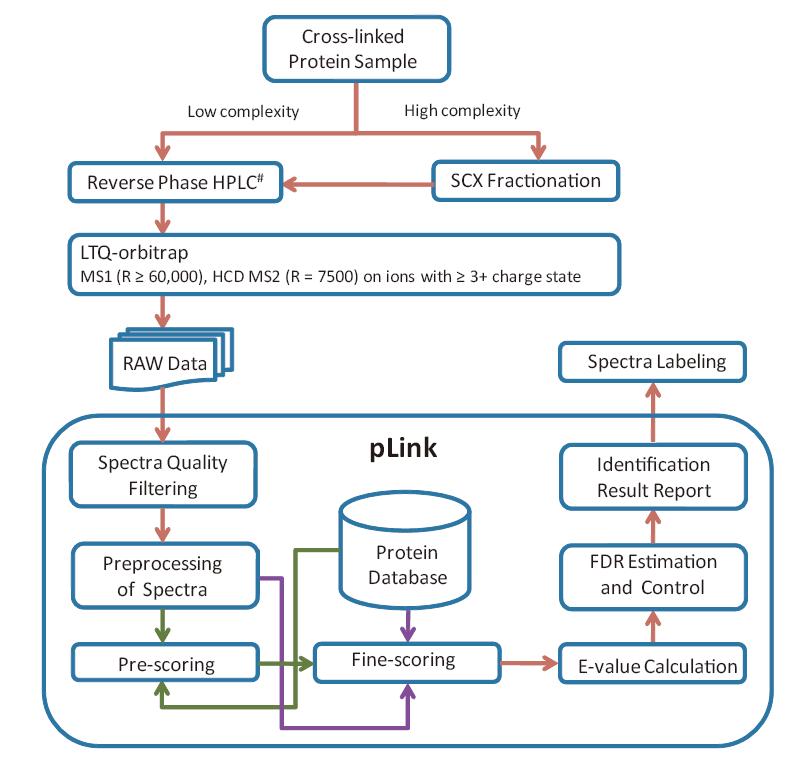
pLink® is a software dedicated for the analysis of chemically cross-linked proteins or protein complexes using mass spectrometry.
For complex samples, pLink can identify cross-linked peptides as well as regular peptides in a single search against large protein databases, providing reliable false discovery rate (FDR) estimations and a quick visualization of the cross-link spectra.
Supplemental Files
pLink 2 is an upgrade version of pLink with higher search speed and a graphical user interface. pLink 2 is recommended for cross-linked data analysis. Download pLink 2.
pLink is currently free to use. Download pLink.
Notice: Please read the pFind Studio Software License Agreement before downloading and using the software. Please fill in the registered table and email it to plink@ict.ac.cn to get the registration key.
pLink contains a quick visualization of the cross-link spectra. For more complex functions on visualization, please use pLabel.
For discussion, please visit here.
* If pLink expired, please redownload the software and reinstall it. The old registration key can still be used. Thanks for the support in the past year!
Release notes
1.23 released!
Update: 2015.8.31
* Release pLink 1.23 for filtering separately in intra and inter-molecule.
Update: 2015.1.27
1.22 released!
* Bug fix:
1 The site miss problem when using linker SS is fixed.
2 Modify.ini is updated to suit the new version pLabel.
3 Language problem is fixed.
Update: 2015.1.4
1.21 released!
Update: 2014.11.4
1.20 released!
* New function:
1 For SUMO identification, the e-value for alpha peptide and beta peptide is also given.
2 The default preprocess algorithm is changed, ETD/CID is now better supported.
* Bug fix:
1 The bug that DSS linker will go through the SS flow has been fixed.
Update: 2014.7.16
1.19 released!
* Bug fix:
1 A bug in the filter part has been fixed.
Update: 2014.5.15
1.18 released!
* Bug fix:
1 Results can be correctly generated now, when the results contain peptide that will be inferred to more than 1000 proteins.
Update: 2014.4.8
1.17 released!
* New function:
1 SUMO search is now supported. Use the following parameter:
linker.name1=SUMO
sumo=1
search_mode=2
sumoseq=VYQEQTGG
fixsite=G
targetsite=K
Will use VYQEQTGG as the sumo sequence for search. And the fixsite mean the AA site on sumoseq, targetsite mean the AA site on the peptide.
Also change the linker name to SUMO.
pLink will use fragment ions from both the main peptide and the sumo sequence peptide, which will improve the identification of sumo modified peptides.
Update: 2013.12.31
Happy New Year!
pLink has been updated for 2014.
For expired registration code, just redownload and the old code still work.
If it is convenient to you, please email to plink@ict.ac.cn to help the authors know who are still using the software.
Thanks for your support in the past year!
Update: 2013.12.16
1.16 released!
* New function:
1 Multithread search is now supported. Use the following parameter:
processor_num=7
Will use seven threads for searching. Notice that this is only supported with large database under open flow, since small database search will not cost too much time and may have problem.
If not set, will use 1 as default.
* Bug Fix:
1 For peptide candidate long than 100, pLink will no longer crash.
2 Fixed modification will no longer be considered in max modification number.
Update: 2013.11.5
1.15 released!
* Candidate peptides missing has been fixed.
* Illegal link site (modify site, cleave site) will be checked now.
* Evalue_max parameter can now work properly.
Update: 2013.10.14
1.14 released!
* FDR control is now supported.
* Search mode can be seleted now.
*** It is strongly recommended that do not use exhaustion mode for large database.
* Bugs found when using MGF from other instruments are now fixed.
Update: 2013.9.18
1.13 released!
* Bugs on 32-bit machine are now fixed.
Update: 2013.8.15
* A result merge bug has been fixed.
* A new setup dialog is used.
* Asymmetric crosslinking is supported correctly now.
Update: 2013.6.13
* MGF files from instruments other than Thermo is supported now.
* Minor modification of the user guide.
* Inter/Loop/Mono type results are now exported correctly.
* Labeling software pLabel are now added to pLink setup kit.
Update: 2013.4.28
* Inter/Loop/Mono type results are clearly separated now.
* For large database search, algorithms are adjusted, which results in a minor speed increase.
* User guide updated with the changed files structure.
* Source code refactoring.
Update: 2013.2.15
* Expire bug fixed.
Update: 2012.12.22
* Preprocess for instruments other than HCD now worked properly.
* Modification site will be correctly appointed now.
* More parameters opened now, e.g. max modify number.
* Some items are modified in the user guide.
Update: 2012.10.9
* Some items are added to the user guide, e.g. max cleavage setting.
* XCalibur raw format can be correctly supported now.
* Some unformatted MS2 file will not cause crash now.
Update: 2012.6.2
* An user guide is now added.
* An extremly long protein name will not cause crash now.
* No illegal xlink result will be shown now.
* File path with space is now supported.
* MGF and mgf files are both recognized now.
Update: 2012.2.8
* Evalue threshold after FDR filter is now available.
Update: 2011.12.21
* Common/Mono/Loop results export are supported
Update: 2011.12.17
* Change the start number of link site from 0 to 1
 ®Copyright © pFind Team · Institute of Computing Technology, Chinese Academy of Sciences, Beijing, China
®Copyright © pFind Team · Institute of Computing Technology, Chinese Academy of Sciences, Beijing, China