pFind Studio: a computational solution for mass spectrometry-based proteomics
iPRG 2015
Study Goal of iPRG 2015: Differential Abundance Analysis in Label-free Quantitative Proteomics. Our results are marked by 'pFind' and 'pQuant'. We identified 4284 proteins and quantified 6 differentially expressed proteins.[see the slides of ABRF...]
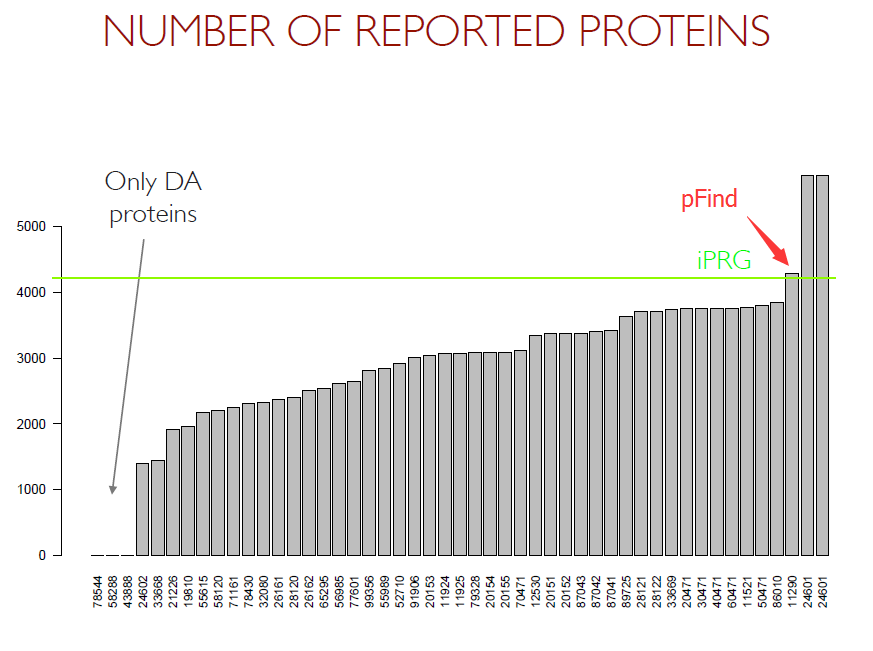

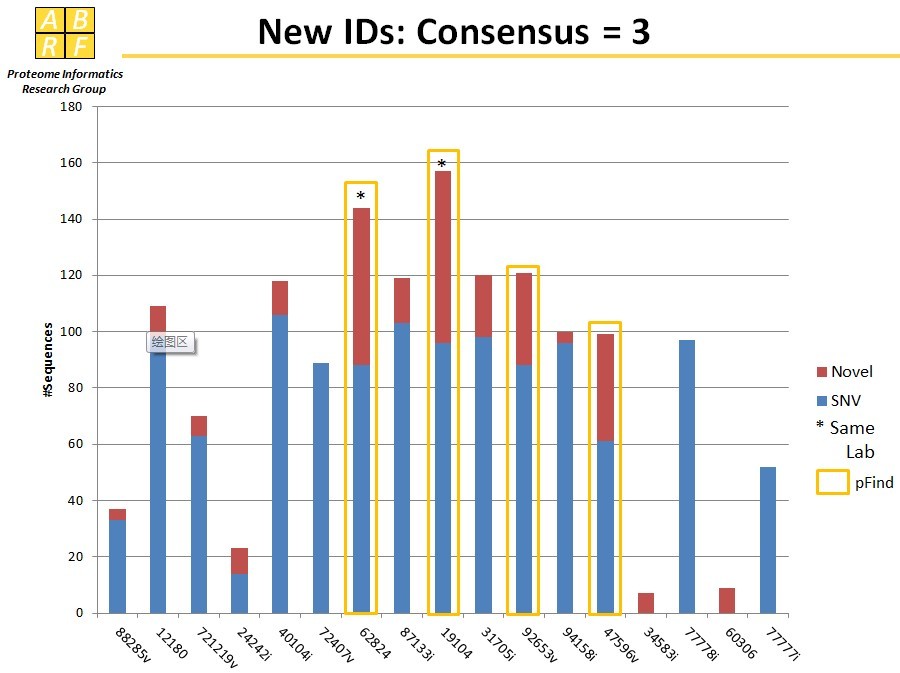
iPRG 2013
Study Goal of iPRG 2013: Compare number of extra identification due to single nucleotide variants vs. novel sequences. Our results are marked by 'pFind'. [see the slides of ABRF...]
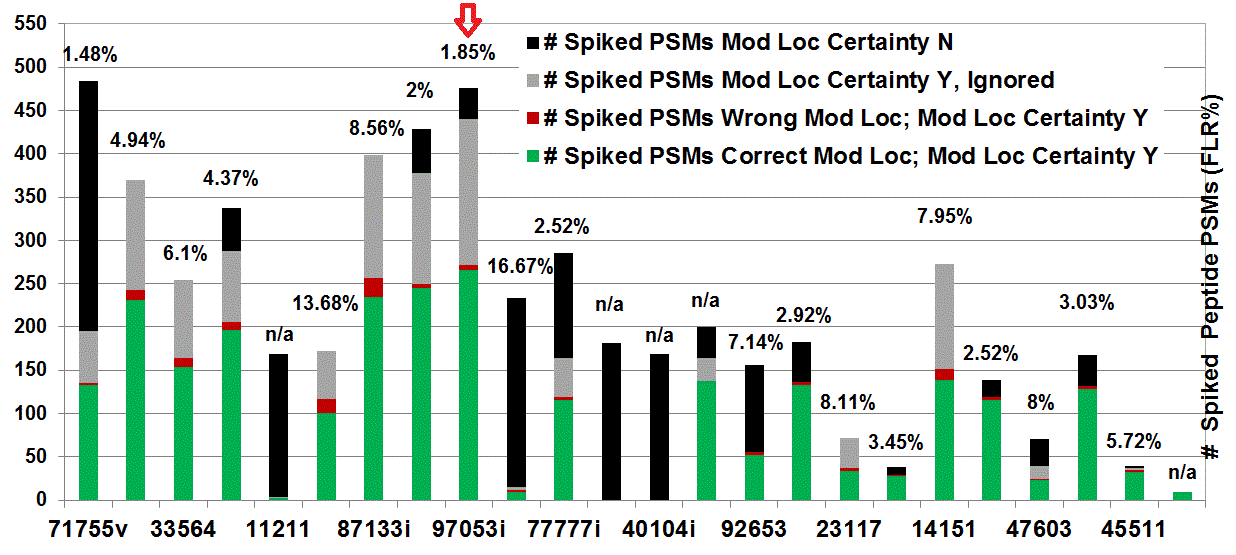
iPRG 2012
This year’s study focused on detecting modified peptides in a complex mixture, with beam-type CID from a Q-TOF type instrument. Our result is numbered ‘97053i’. [see the poster of ABRF...]
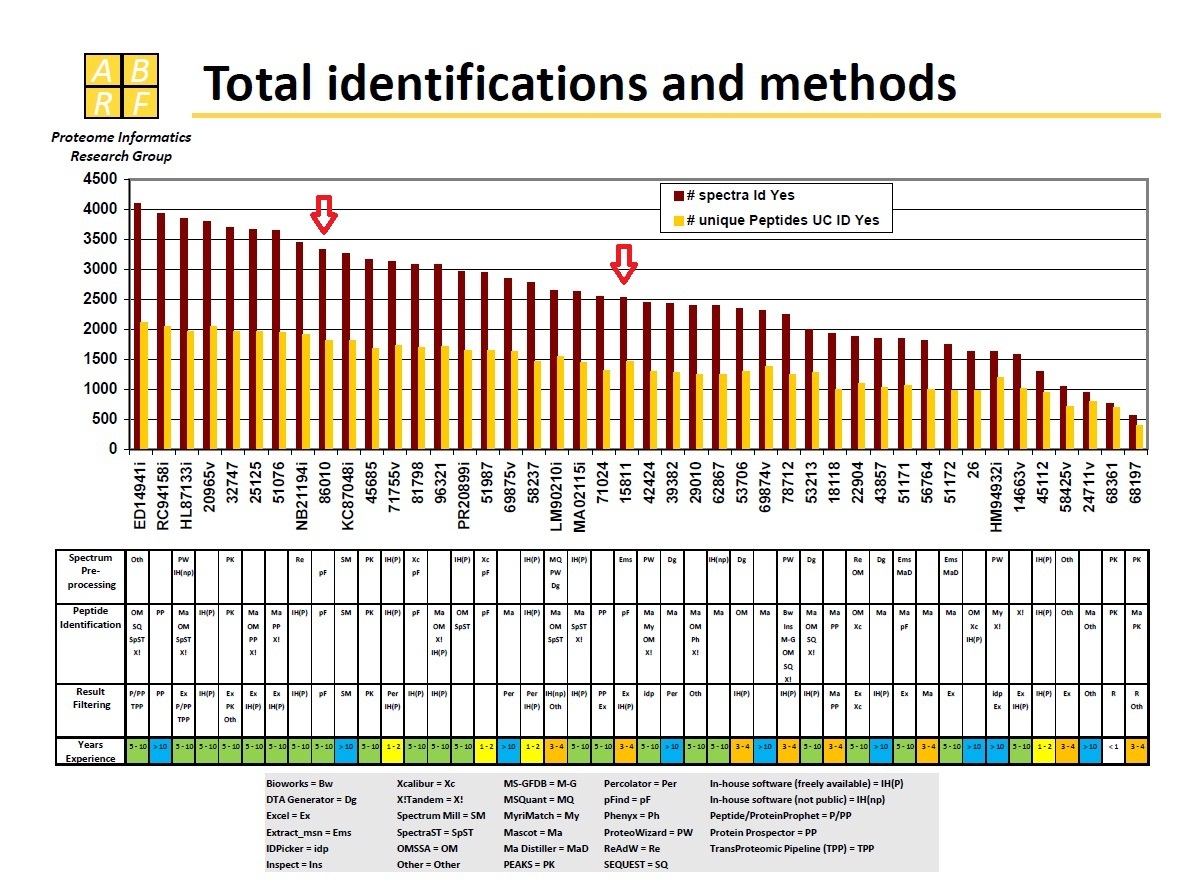
iPRG 2011
iPRG 2011: A Study on the Identification of Electron Transfer Dissociation (ETD) Mass Spectra. We had two submissions numbered ‘86010’ and ‘15811’. [see the slides of ABRF...]
iPRG 2010
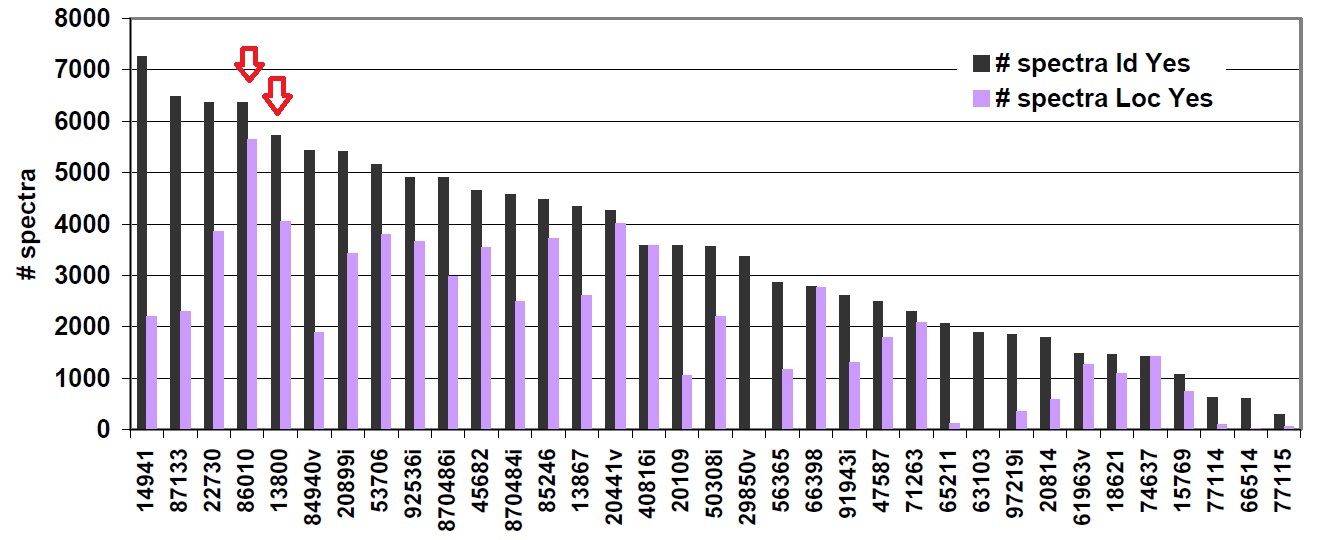
In January 2010, we participated the ABRF iPRG study with pFind, developed by our group. This year’s study focused on the evaluation to identify the phosphopeptides and localize the phosphorylation sites. In the study, an LC-MS/MS dataset from a lysate digested with trypsin and enriched for phosphopeptides using strong cation exchange fractionation followed by immobilized metal affinity chromatography (SCX/IMAC) was provided. This study helped the participants to evaluate their data analysis capabilities and approaches relative to others in analyzing a common data set. We had two submissions numbered ‘86010’ and ‘13800’, For more details, please refer to the following documents or ABRF website.
1. iPRG 2010 study announcement
2. iPRG 2010 presentation slides
iPRG 2008
In January 2008, we participated the ABRF iPRG study with pFind (a database-searching software for protein identification via tandem mass spectrometry) and pCompare (an in-house software tool for peptide identification result parsing and protein inferring) developed by ourselves. The primary goal of this study is to evaluate the current levels of reporting protein identification results in different labs. According to the independent evaluation by ABRF iPRG, our software achieve comparative performance to that of experts in iPRG on an average with a relatively lower false discovery rate. Now we are taking our efforts to make pFind more reliable and applicable. [more...]
KDD-Cup
KDD Cup is the worldwide Data Mining and Knowledge Discovery competition organized by ACM SIGKDD (Special Interest Group on Knowledge Discovery and Data Mining). KDD Cup is held annually in conjunction with the ACM SIGKDD conferences. It aims at showing the best methods for discovering higher-level knowledge from data, helping to close the gap between research and industry and stimulating further KDD research and development. [more...]